Output files
The output of the analyses performed with oncoEnrichR are provided in two different formats:
- An interactive quarto-generated HTML report
- A multi-sheet Excel workbook
HTML report
Here, we showcase screenshots from the oncoEnrichR interactive HTML report, following the various types of questions that can be answered with the tool.
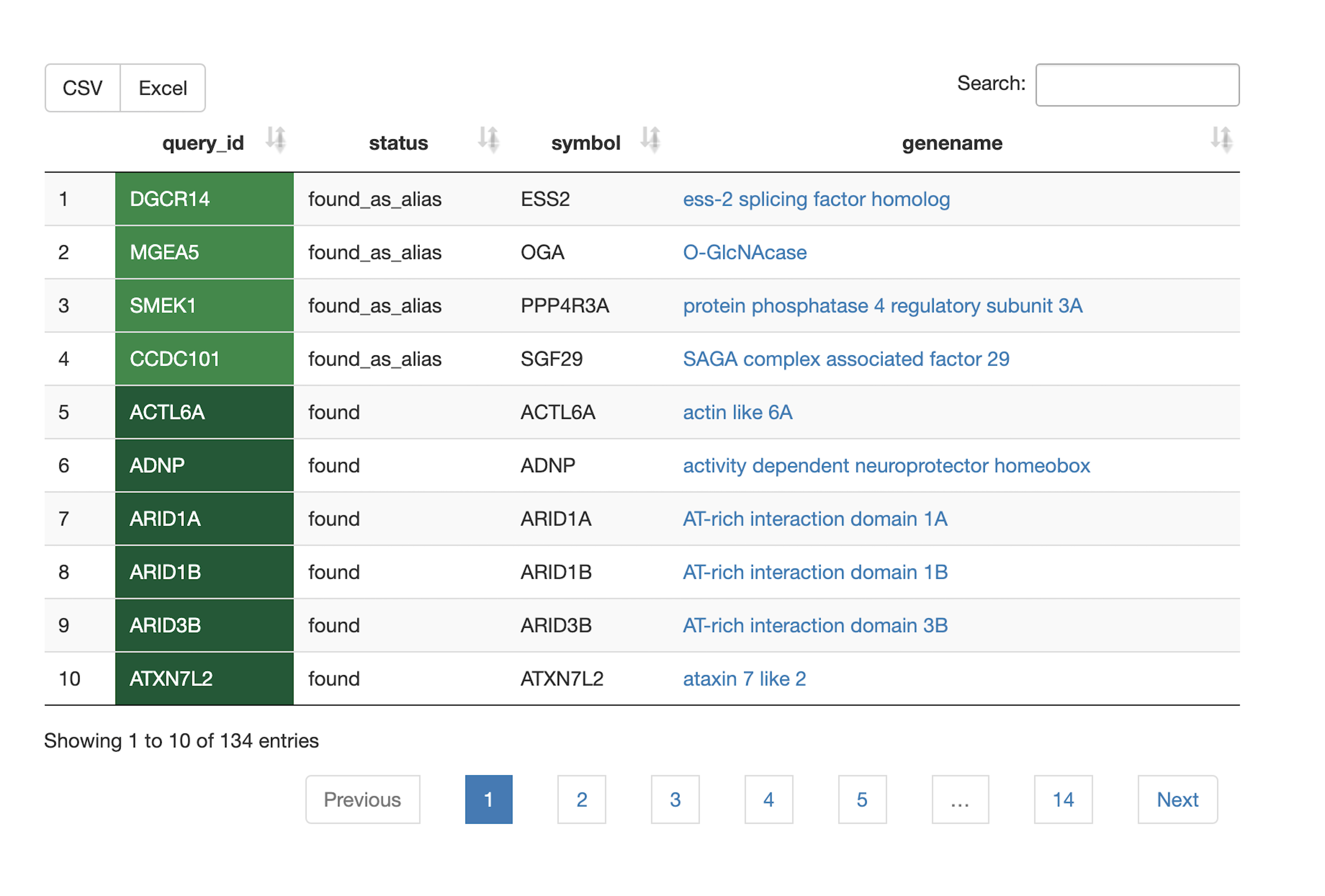
Query verification
- Are all my gene/query identifiers correctly identified? Are some of my query identifiers outdated, and no longer considered the primary identifier?
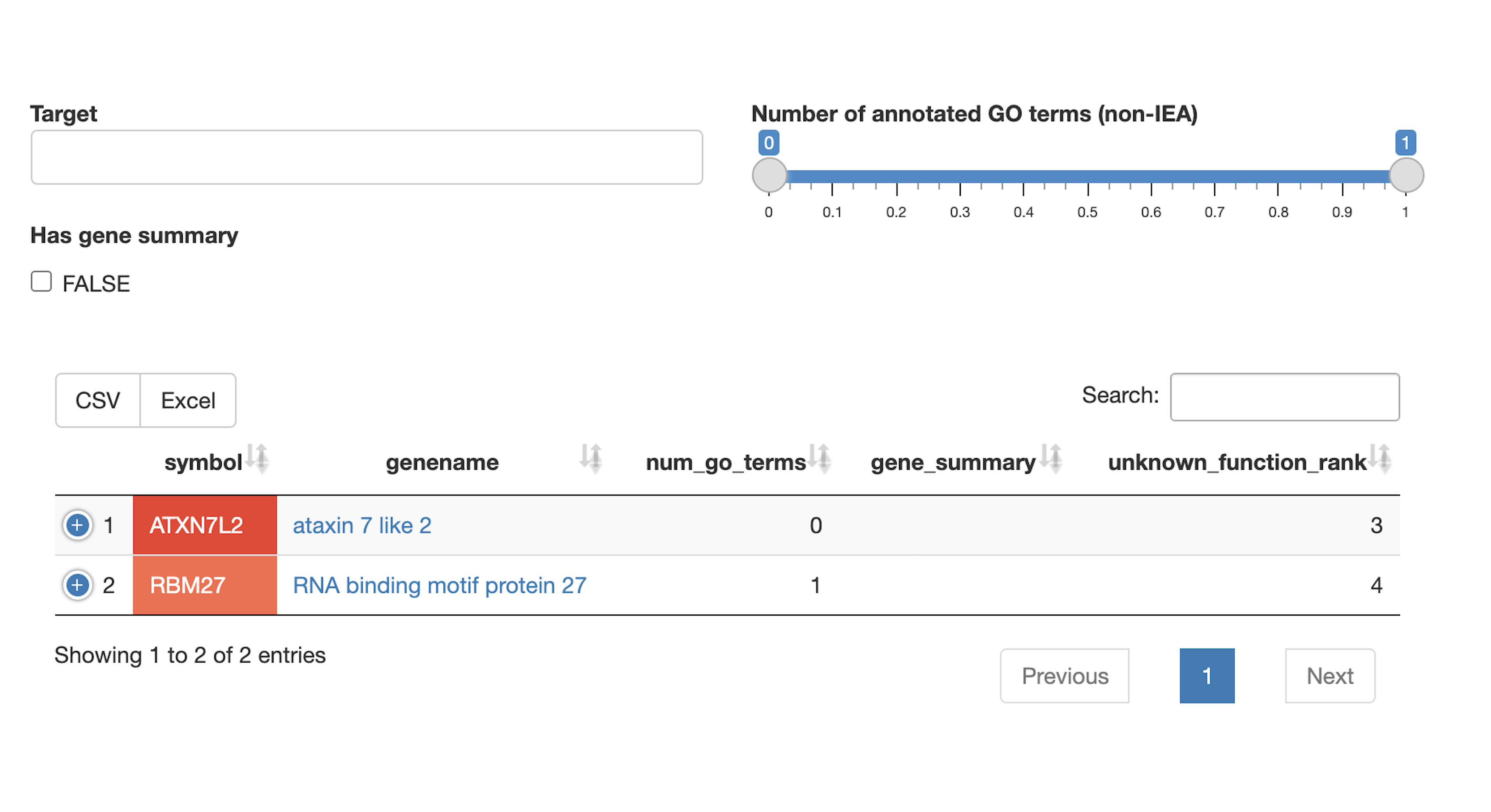
Poorly characterized genes
- Which members of the query set have a poorly characterized/unknown function?
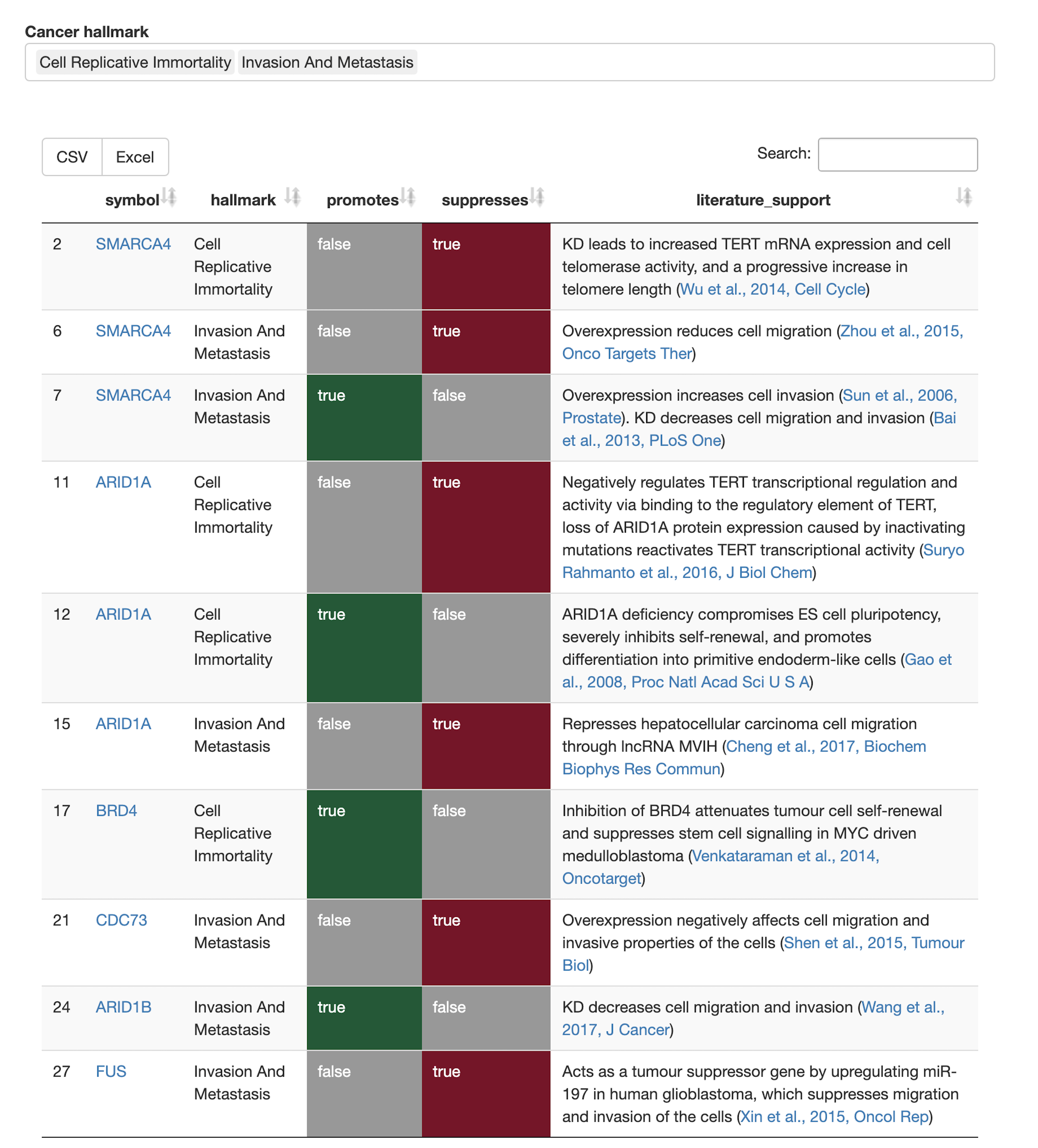
Cancer associations
- Which tumor types are associated with genes in the queryset? To what extents? Which genes are classified as tumor suppressors, proto-oncogenes, or potential cancer driver genes?
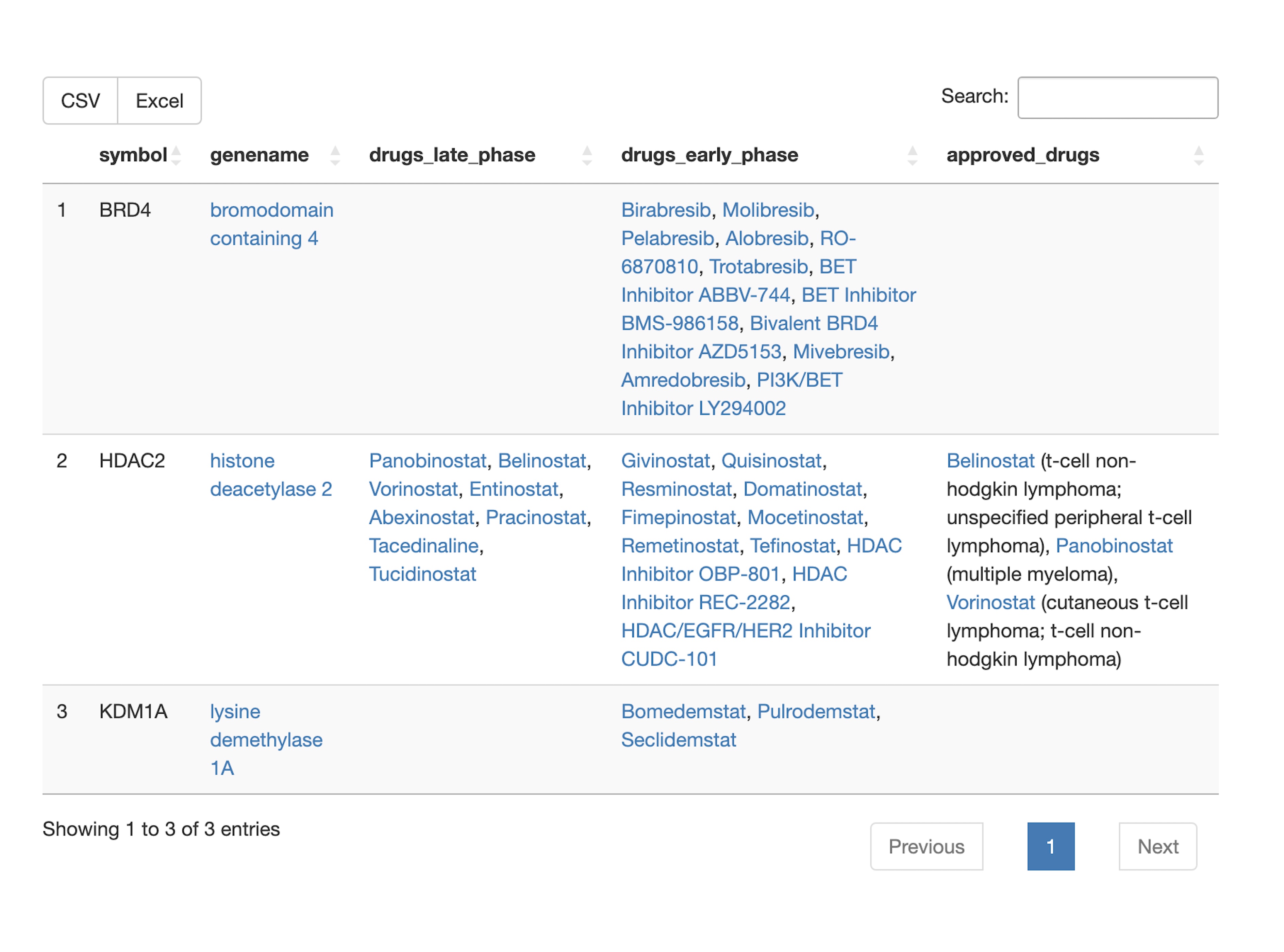
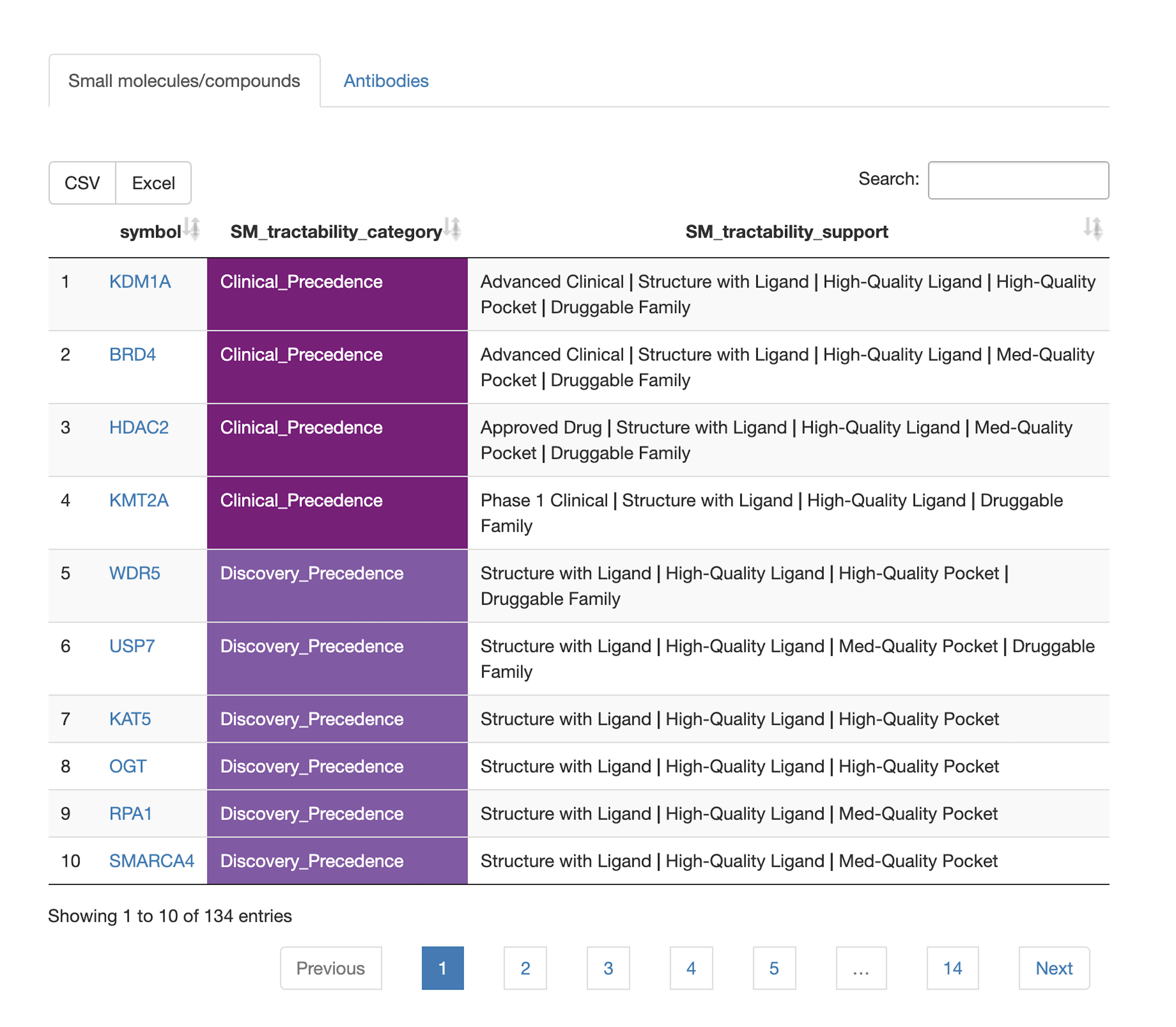
Drug associations
- Are there available targeted cancer drugs for query set members? For which cancer indications are there approved drugs targeted towards query set members? What are the target tractabilities of all query set members?
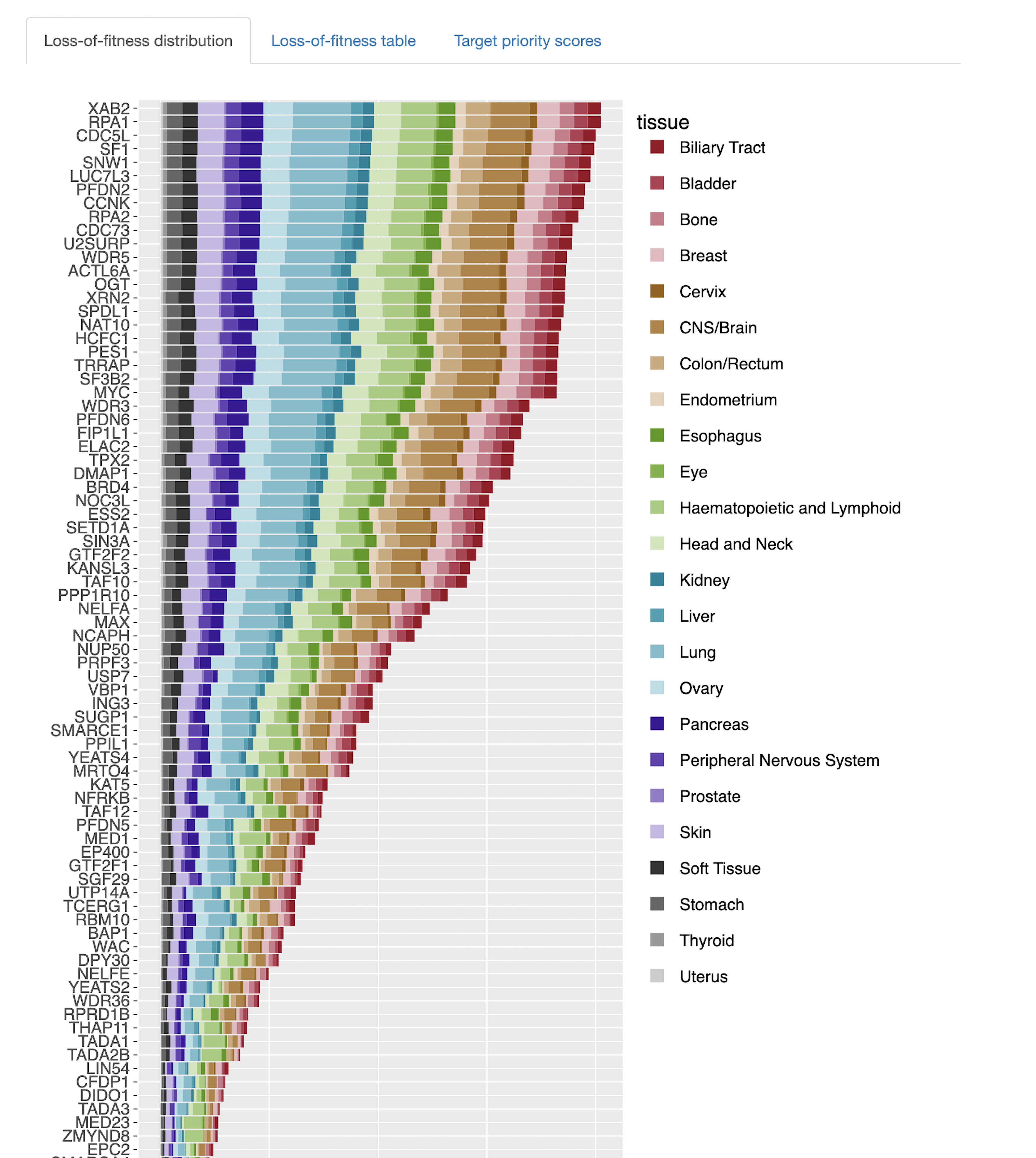
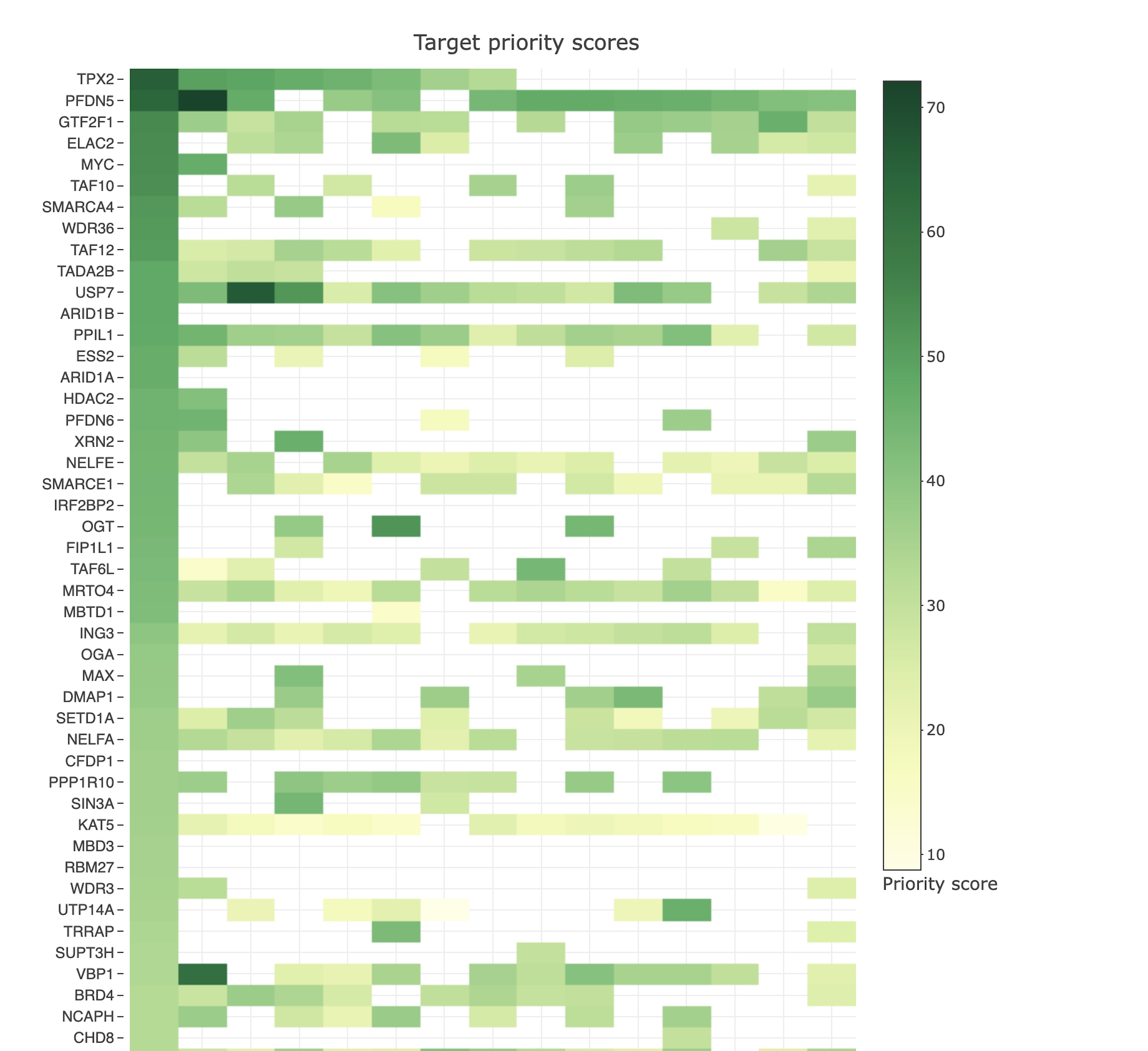
Gene fitness scores
- Which members in the query set have a significant effect on cell fitness, considering large-scale genome-wide CRISPR–Cas9 dropout screening data of cancer cell lines?
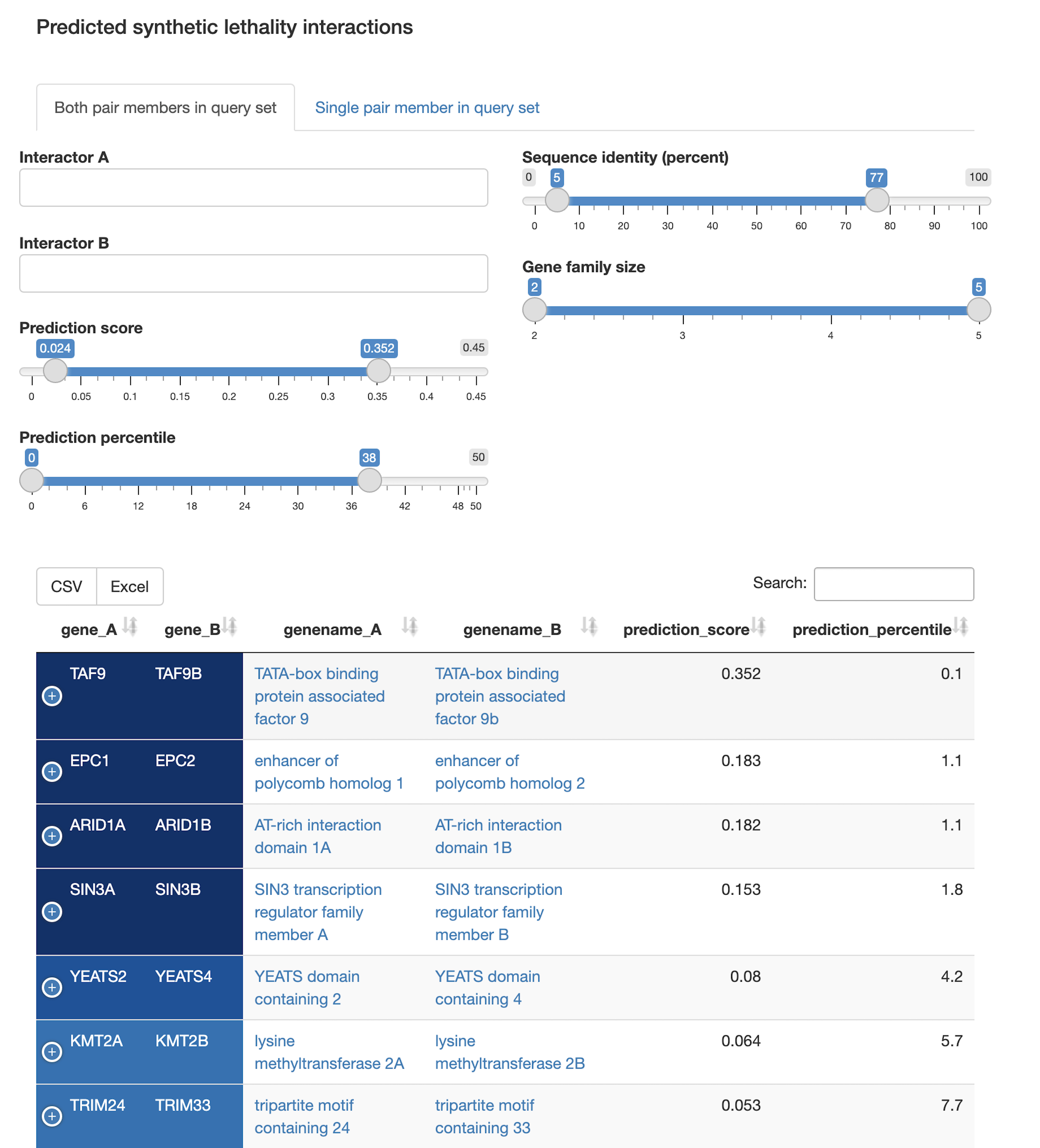
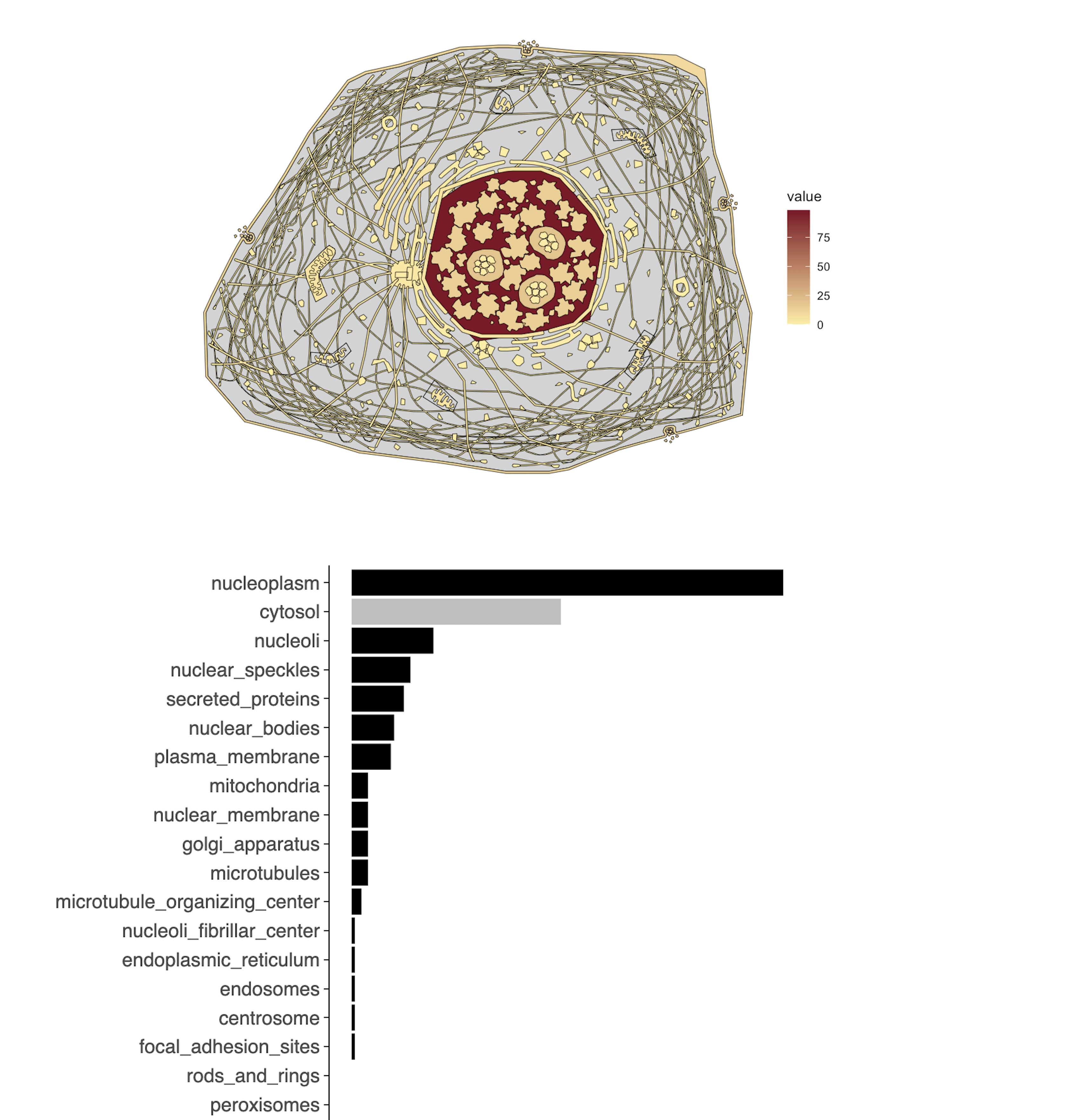
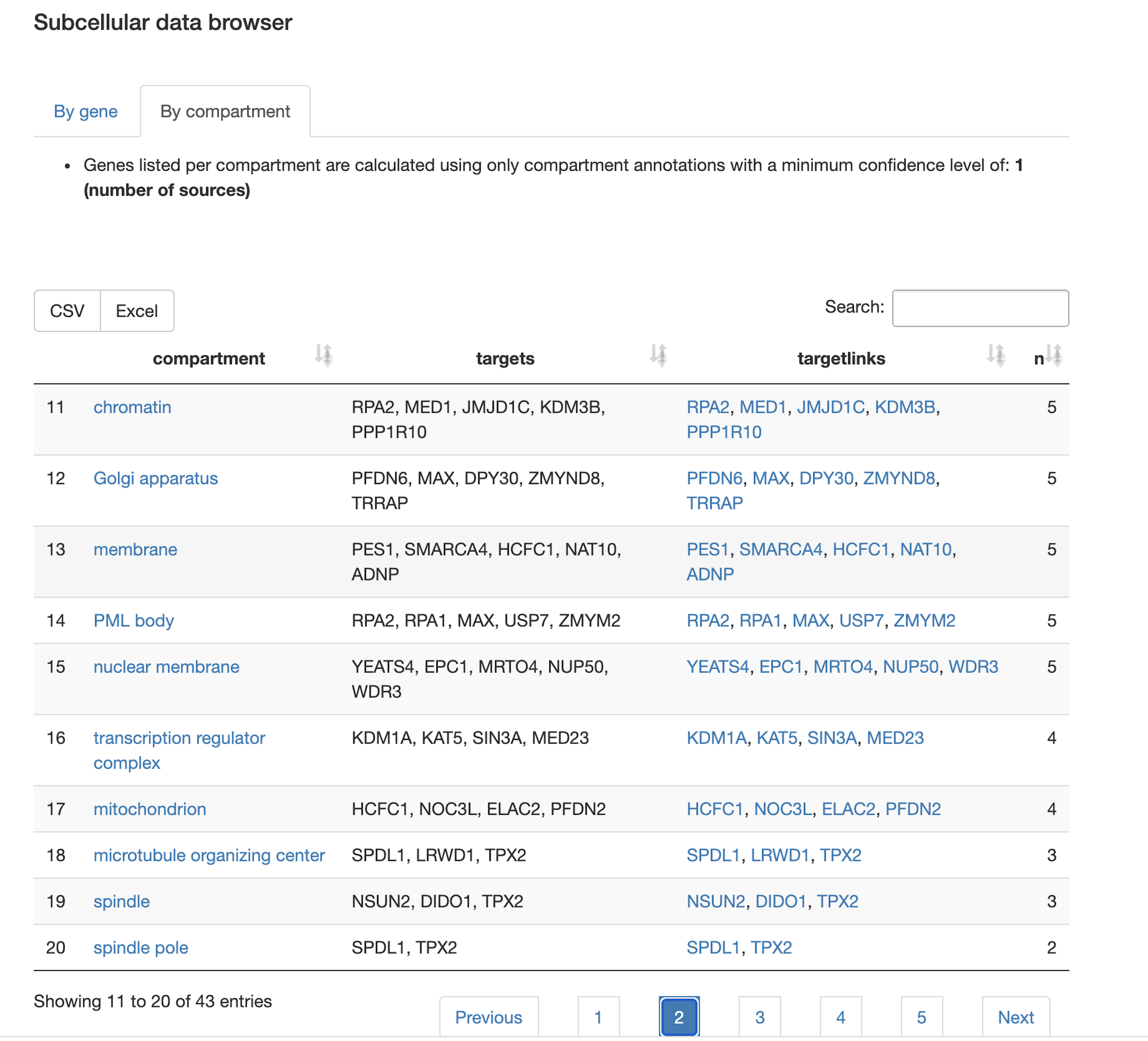
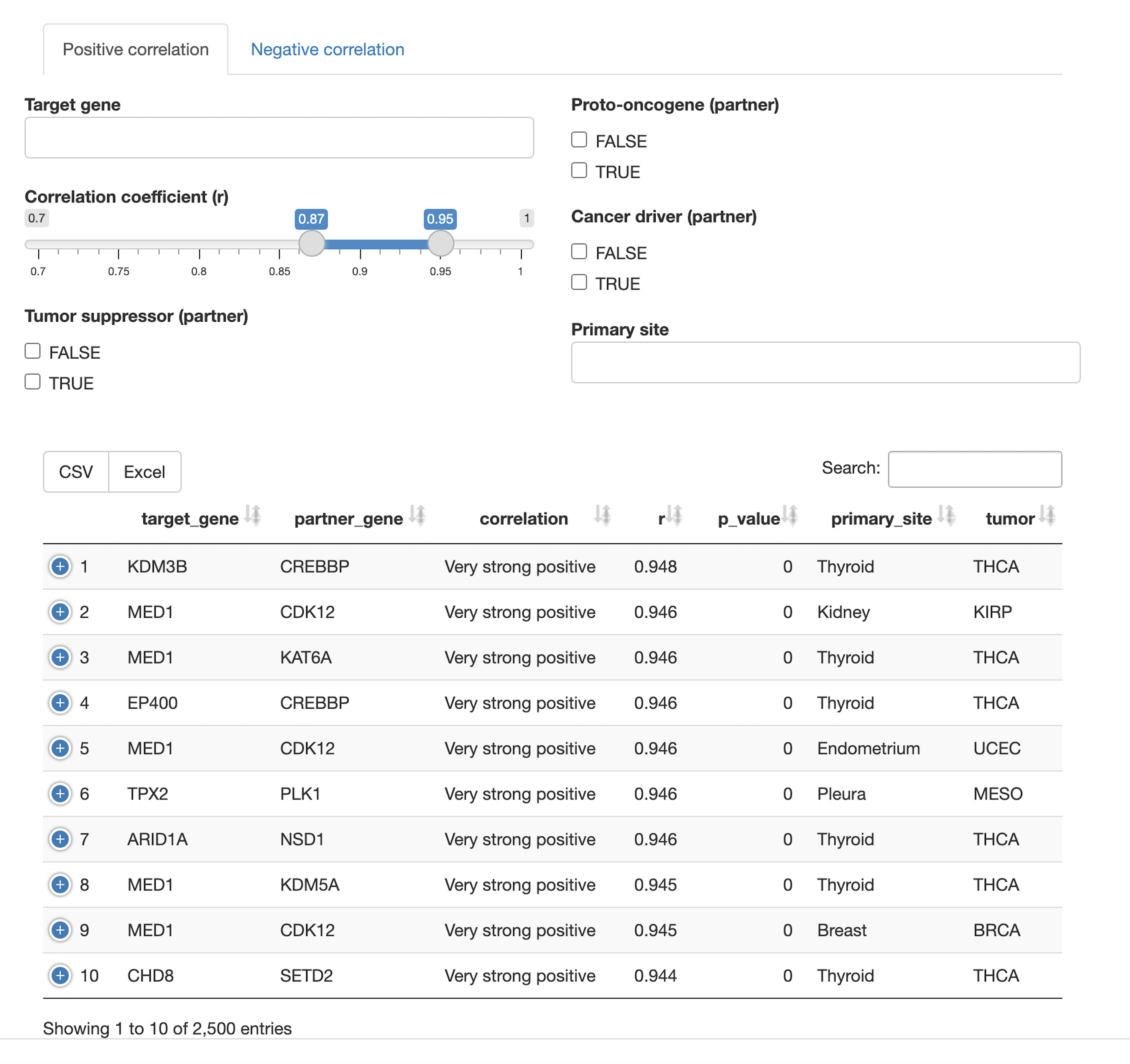
Protein-protein interaction network
- How are protein-protein interactions represented in the query set? Which proteins are hubs in the interaction network? Which members of the query set form community structures in the network?
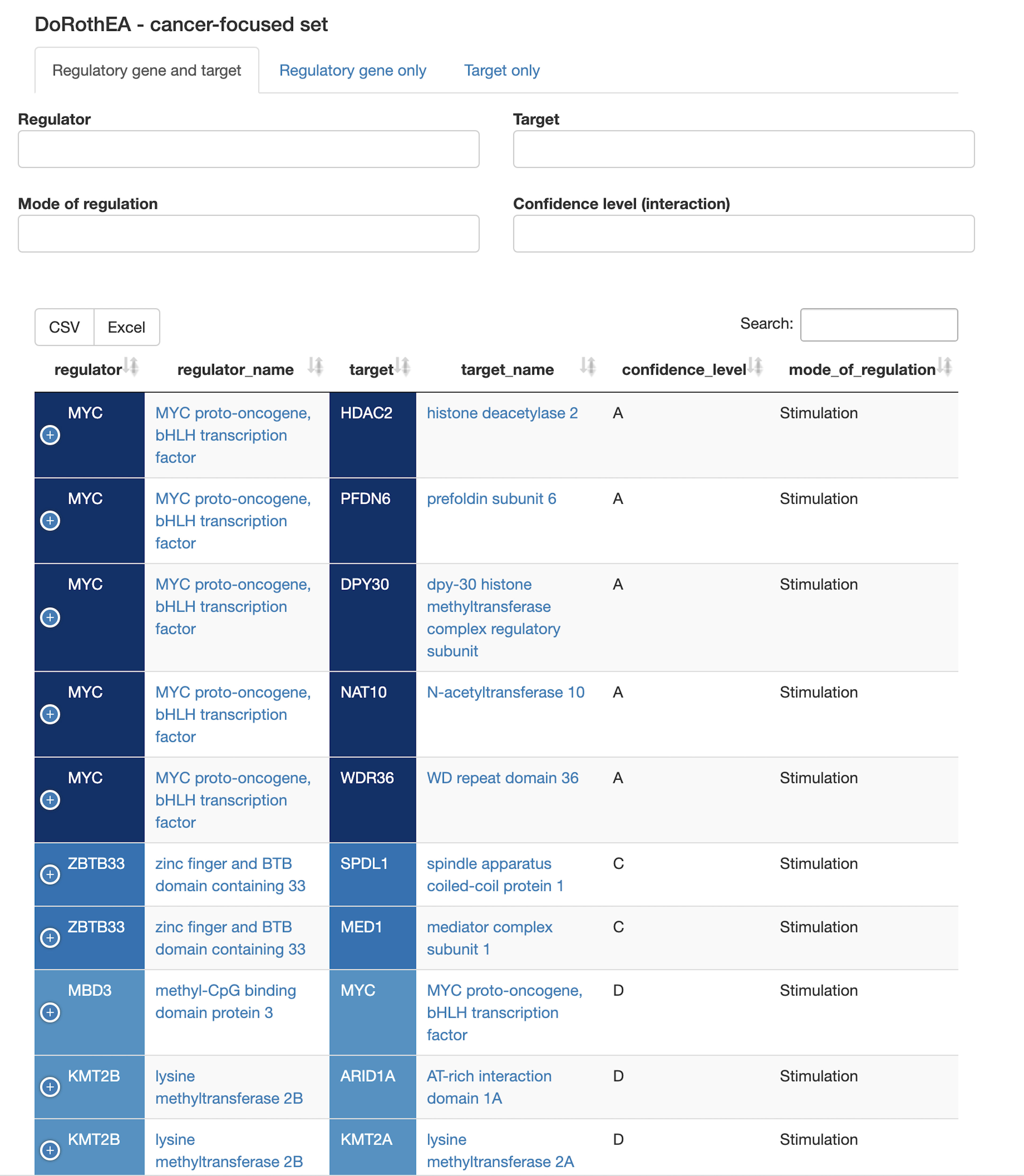
Regulatory interactions
- Are there transcriptional regulatory interactions in the query set? Both TF/regulator and target present? What are the regulatory targets for other TFs present? What are the regulators of other targets?
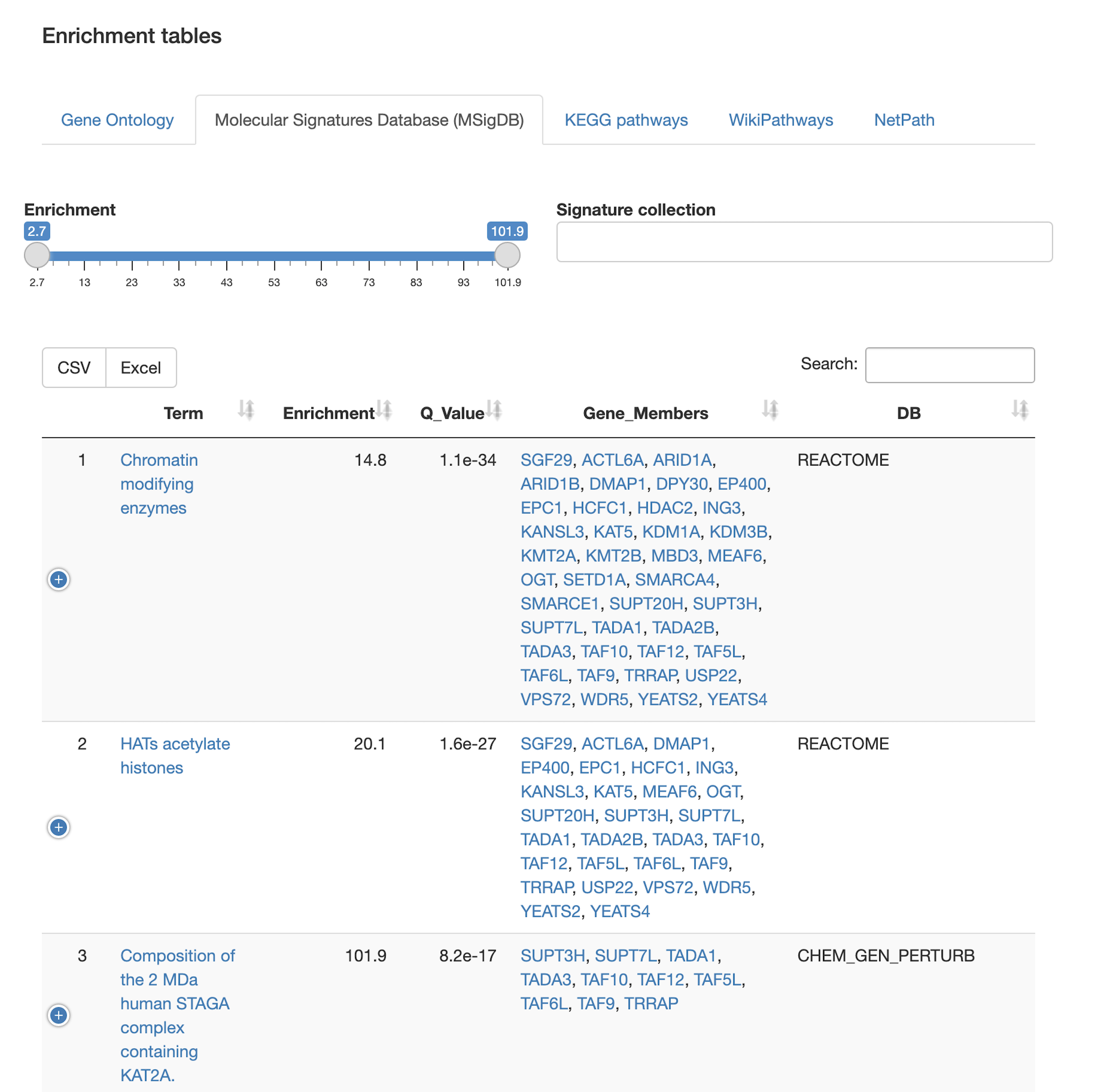
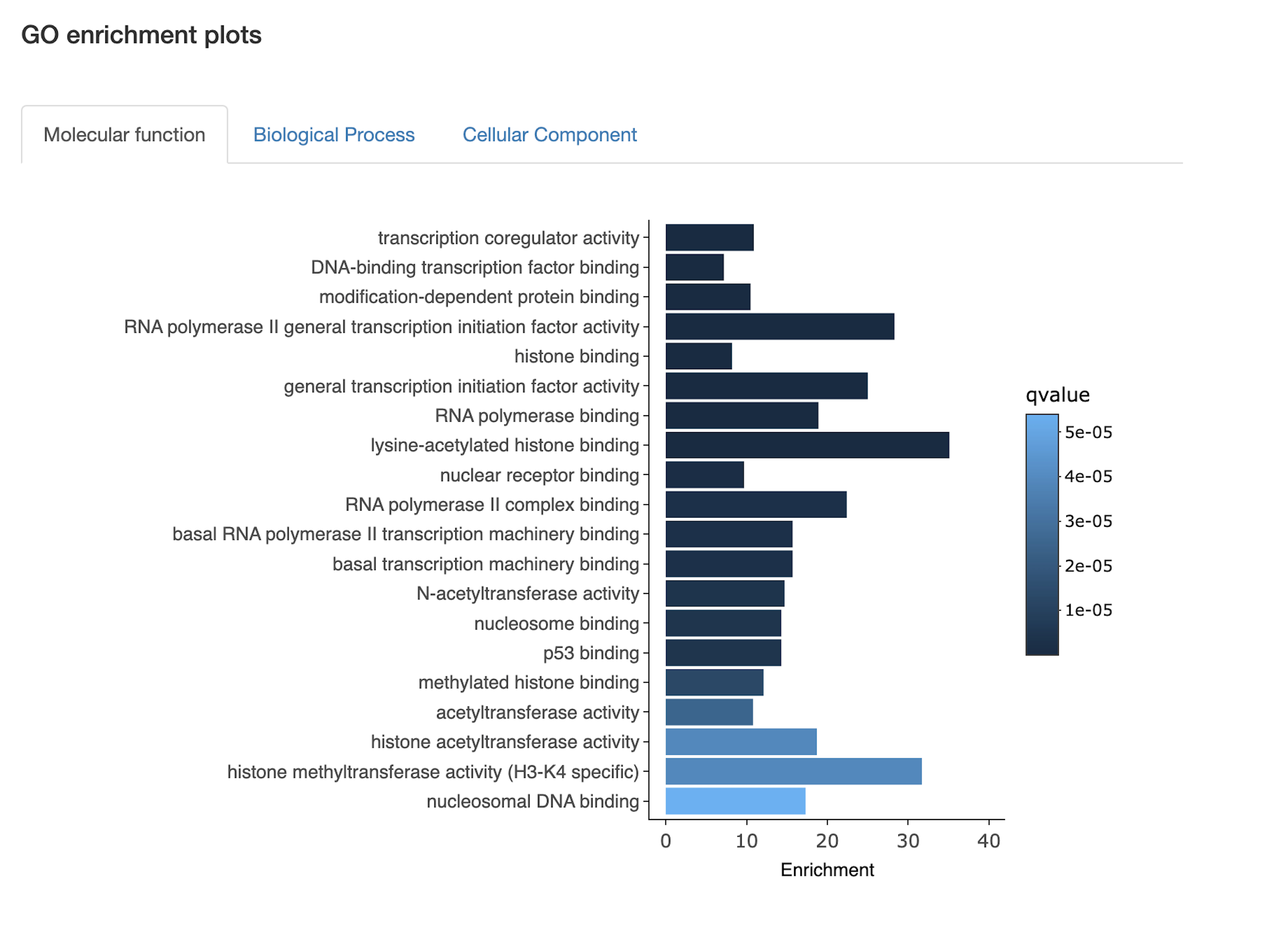
Function and pathway enrichment
- Which biological pathways/processes, molecular functions, or cancer gene signatures are enriched in the query set?
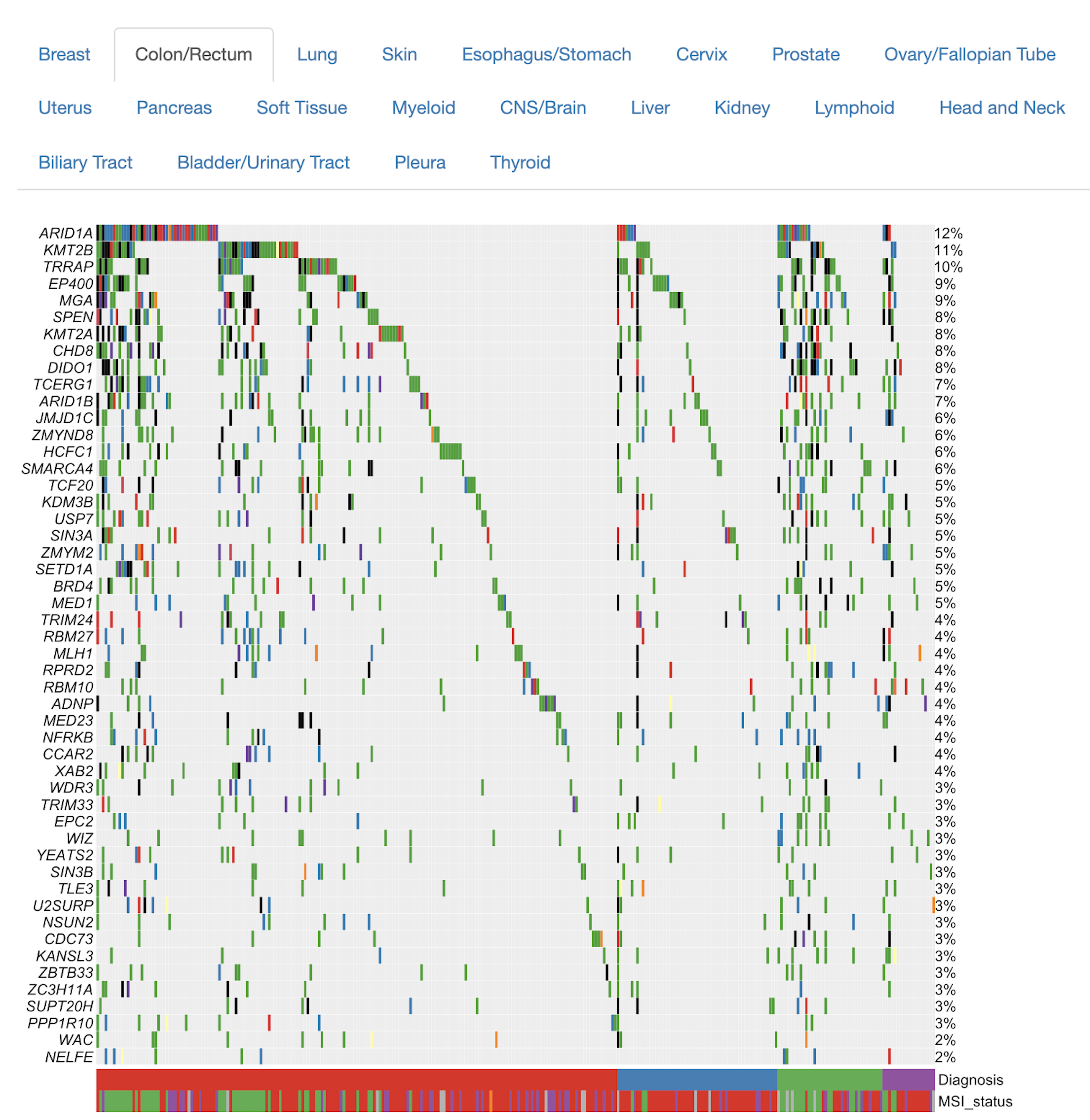
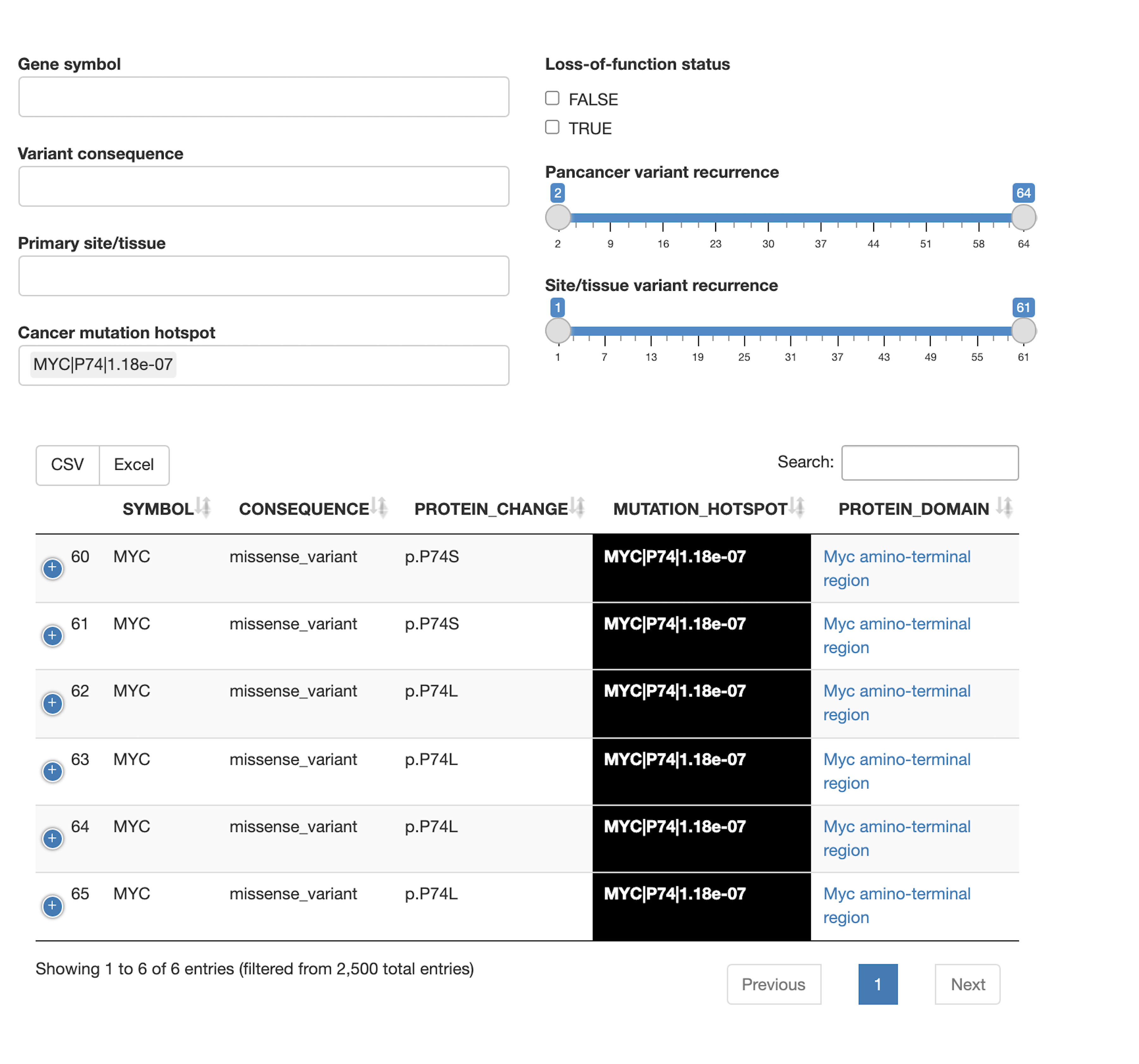
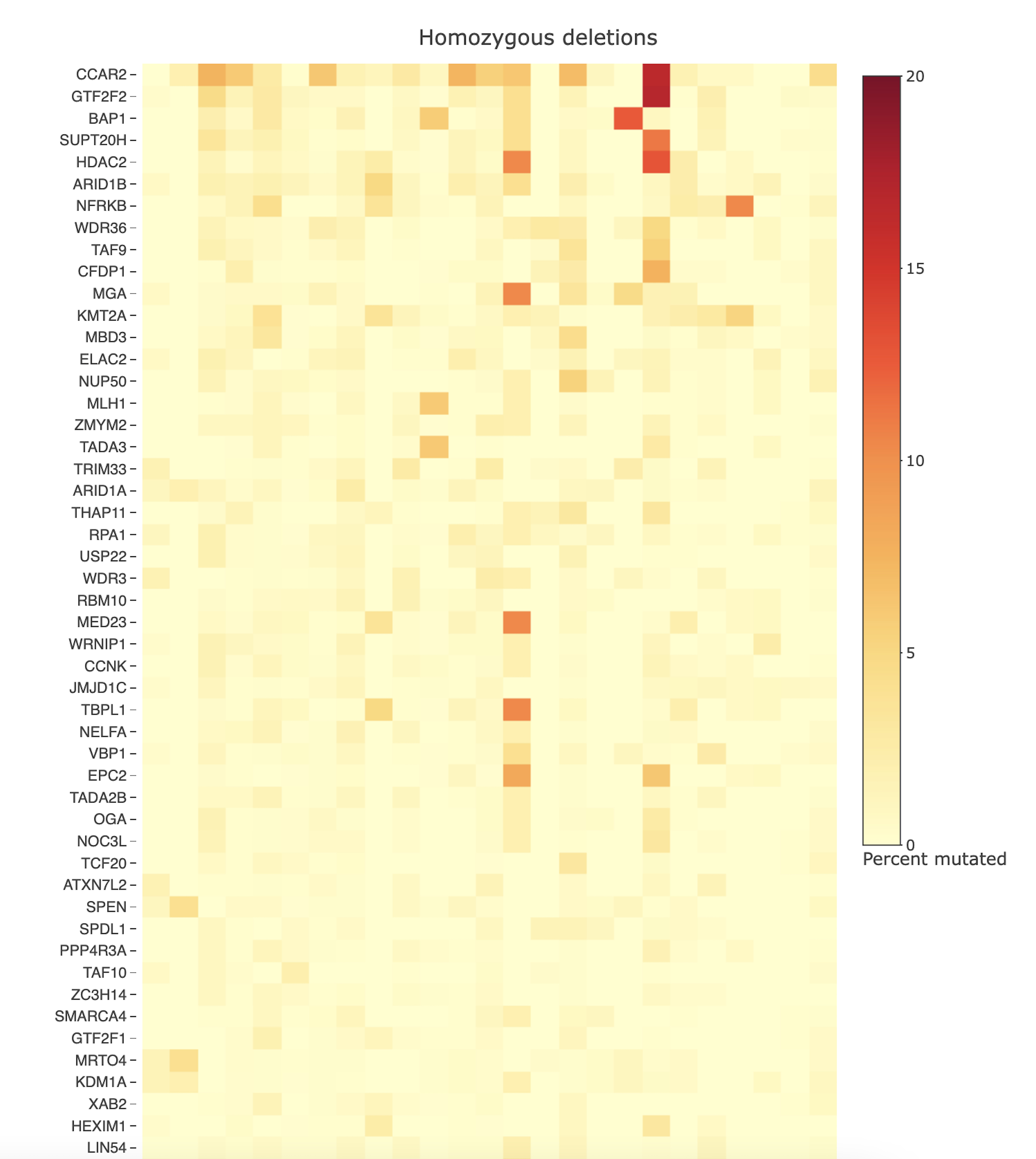
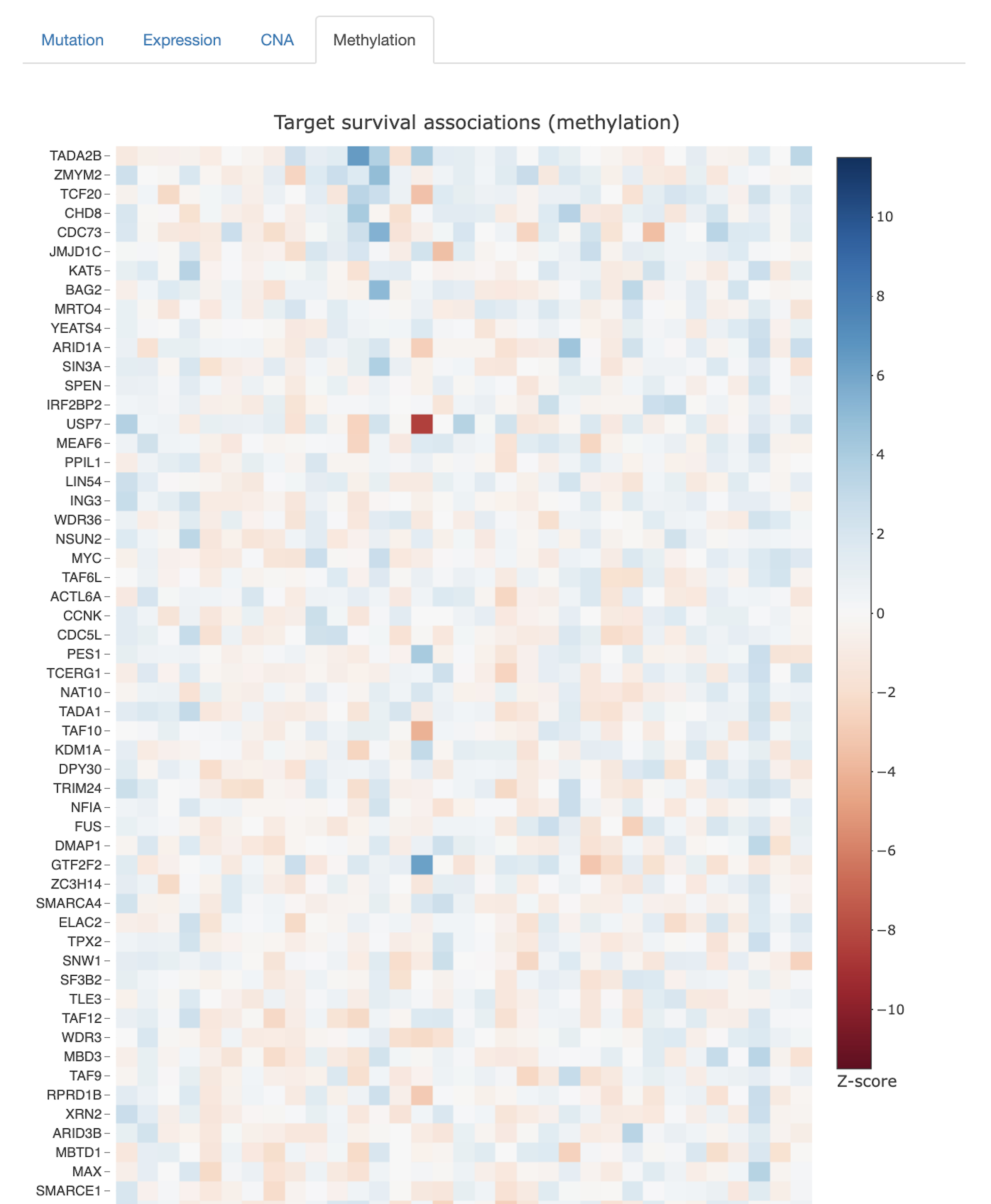
Tumor aberration frequencies
- To what extent are genes in the query set mutated in tumor samples, through copy number alterations or point mutations/indels?